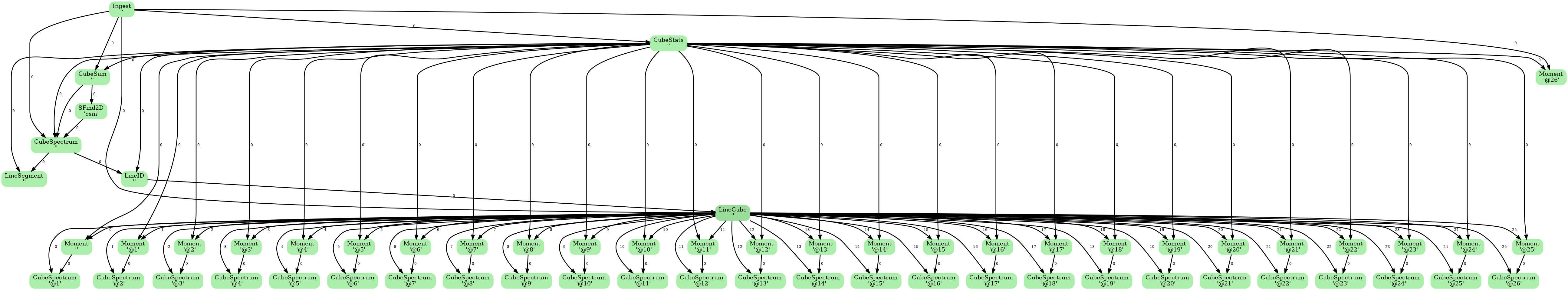
This diagram is a directed acyclic graph representing the ADMIT Task connections for the flow. Each arrow represents the connection from an ADMIT task output to the input of another ADMIT Task. The integer next to each arrow is the zero-based index of the ADMIT Task's output BDP. Note any output BDP may be used as the input for more than one ADMIT Task.
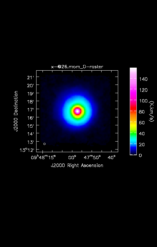
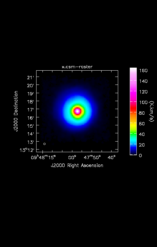
Ingest_AT reads in a FITS image into ADMIT, creating a CASA image and extracting basic information. It is usually the first step in any ADMIT flow.
| Input image (FITS or CASA) | /lma1/teuben/lmtoy/examples/2018S1SEQUOIACommissioning/79448/IRC+10216_79448.nfs.fits |
| Output image (CASA) | x.im |
| Object | IRC+10216 |
| Rest frequency | 115.271204 GHz |
| VLSR | -26.0 km/s (estimated) |
| Beam | 14.38" x 14.38" PA @ 0.00 deg |
| Imsize | 97 x 97 x 689 |
| Intensity unit | K |
| Data min/max | -7.142E-02 / 6.078E+00 |
| Telescope | LMT |
| Percentage bad pixels | 6.74% |
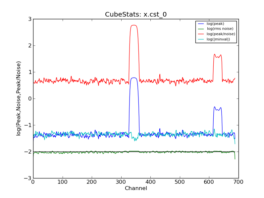
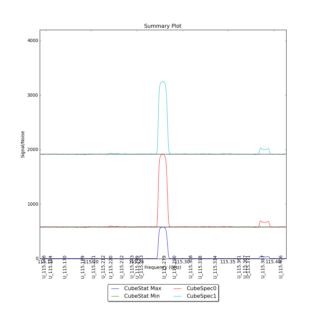
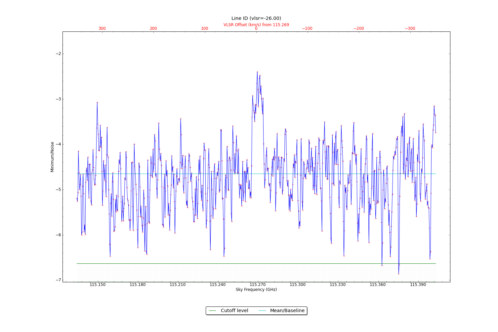
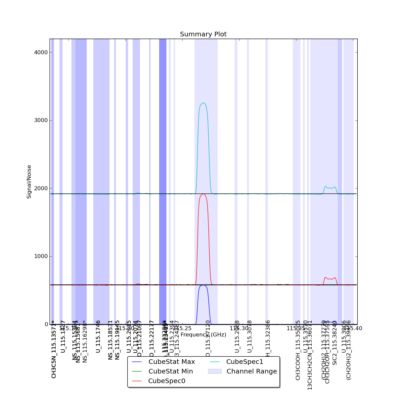
CubeStats_AT computes image-plane robust statistics on datacubes. These statistics are particularly useful for identifying spectral lines in images where the noise varies as a funnction of frequency.
| CASA image | x.im |
| RMS method | medabsdevmed |
| RMS value | 9.881E-03 |
| Dynamic range | 6.151E+02 |
| Data mean | 7.370E-03 |
CubeSum_AT sums all the emission in a cube, regardless of whether or not it comes from different spectral lines, creating a integrated map of all emission.
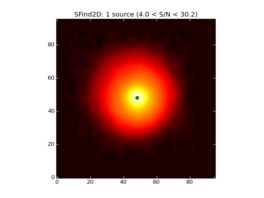
SFind2D_AT creates a list of sources found in a 2D image based on the noise statistics in the image as determined by CubeStats.
CubeSpectrum_AT computes spectra at one or more (x,y) positions in the datacube. Alternatively, it can produce a spectrum averaged over a box.
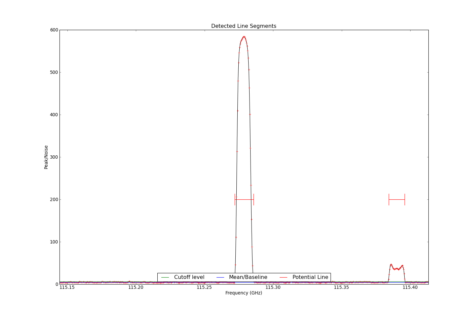
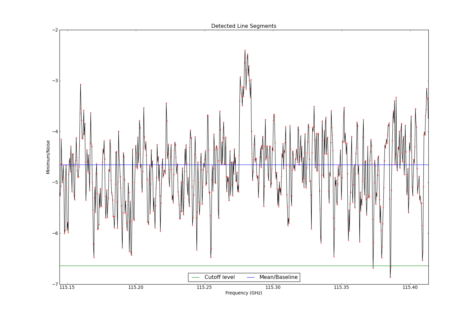
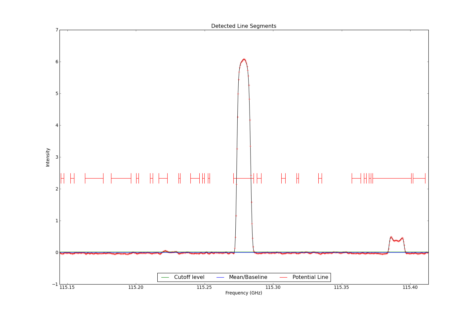
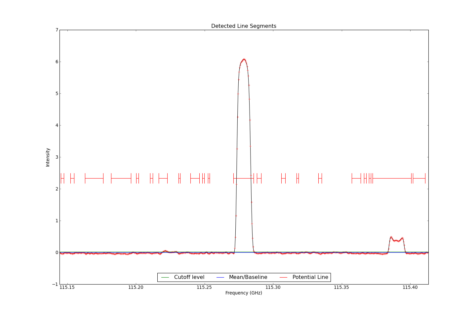
LineSegment_AT identifies regions in one or more spectrum that have contiguous emission above a user specified noise floor. The input spectra are CubeSpectrum_AT or CubeStats_AT.
Line Segments
| frequency | uid | formula | name | transition | velocity | El | Eu | linestrength | peakintensity | peakoffset | fwhm | startchan | endchan | peakrms | blend | force | [GHz] | [] | [] | [] | [] | [km/s] | [K] | [K] | [D^2] | [Jy/beam] | [km/s] | [km/s] | [] | [] | [] | [] | [] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1.151E+02 | U_115.146 | NotIdentified | Not Identified | 0.000E+00 | 0.000E+00 | 0.000E+00 | 0.000E+00 | 7.952E-01 | 0.000E+00 | 5.592E+00 | 2.000E+00 | 9.000E+00 | 2.455E+00 | 0.000E+00 | False | |
| 1.152E+02 | U_115.154 | NotIdentified | Not Identified | 0.000E+00 | 0.000E+00 | 0.000E+00 | 0.000E+00 | 1.163E+00 | 0.000E+00 | 6.230E+00 | 2.000E+01 | 2.800E+01 | 3.589E+00 | 0.000E+00 | False | |
| 1.152E+02 | U_115.170 | NotIdentified | Not Identified | 0.000E+00 | 0.000E+00 | 0.000E+00 | 0.000E+00 | 1.826E+00 | 0.000E+00 | 2.599E+01 | 4.700E+01 | 8.200E+01 | 4.599E+00 | 0.000E+00 | False | |
| 1.152E+02 | U_115.189 | NotIdentified | Not Identified | 0.000E+00 | 0.000E+00 | 0.000E+00 | 0.000E+00 | 2.492E+00 | 0.000E+00 | 3.924E+01 | 9.600E+01 | 1.340E+02 | 7.693E+00 | 0.000E+00 | False | |
| 1.152E+02 | U_115.201 | NotIdentified | Not Identified | 0.000E+00 | 0.000E+00 | 0.000E+00 | 0.000E+00 | 5.394E-01 | 0.000E+00 | 4.389E+00 | 1.430E+02 | 1.480E+02 | 1.359E+00 | 0.000E+00 | False | |
| 1.152E+02 | U_115.212 | NotIdentified | Not Identified | 0.000E+00 | 0.000E+00 | 0.000E+00 | 0.000E+00 | 9.577E-01 | 0.000E+00 | 9.613E+00 | 1.690E+02 | 1.750E+02 | 2.412E+00 | 0.000E+00 | False | |
| 1.152E+02 | U_115.220 | NotIdentified | Not Identified | 0.000E+00 | 0.000E+00 | 0.000E+00 | 0.000E+00 | 1.310E+00 | 0.000E+00 | 1.893E+01 | 1.850E+02 | 2.020E+02 | 3.300E+00 | 0.000E+00 | False | |
| 1.152E+02 | U_115.232 | NotIdentified | Not Identified | 0.000E+00 | 0.000E+00 | 0.000E+00 | 0.000E+00 | 1.435E+00 | 0.000E+00 | 3.155E+00 | 2.220E+02 | 2.260E+02 | 4.429E+00 | 0.000E+00 | False | |
| 1.152E+02 | U_115.243 | NotIdentified | Not Identified | 0.000E+00 | 0.000E+00 | 0.000E+00 | 0.000E+00 | 1.678E+00 | 0.000E+00 | 1.750E+01 | 2.440E+02 | 2.620E+02 | 4.227E+00 | 0.000E+00 | False | |
| 1.152E+02 | U_115.249 | NotIdentified | Not Identified | 0.000E+00 | 0.000E+00 | 0.000E+00 | 0.000E+00 | 1.004E+00 | 0.000E+00 | 4.619E+00 | 2.660E+02 | 2.710E+02 | 3.098E+00 | 0.000E+00 | False | |
| 1.153E+02 | U_115.253 | NotIdentified | Not Identified | 0.000E+00 | 0.000E+00 | 0.000E+00 | 0.000E+00 | 9.500E-01 | 0.000E+00 | 5.924E+00 | 2.770E+02 | 2.810E+02 | 2.393E+00 | 0.000E+00 | False | |
| 1.153E+02 | U_115.279 | NotIdentified | Not Identified | 0.000E+00 | 0.000E+00 | 0.000E+00 | 0.000E+00 | 5.796E+02 | 0.000E+00 | 2.308E+01 | 3.240E+02 | 3.630E+02 | 1.789E+03 | 0.000E+00 | False | |
| 1.153E+02 | U_115.290 | NotIdentified | Not Identified | 0.000E+00 | 0.000E+00 | 0.000E+00 | 0.000E+00 | 6.367E-01 | 0.000E+00 | 8.788E+00 | 3.680E+02 | 3.770E+02 | 1.604E+00 | 0.000E+00 | False | |
| 1.153E+02 | U_115.308 | NotIdentified | Not Identified | 0.000E+00 | 0.000E+00 | 0.000E+00 | 0.000E+00 | 9.391E-01 | 0.000E+00 | 8.682E+00 | 4.140E+02 | 4.220E+02 | 2.899E+00 | 0.000E+00 | False | |
| 1.153E+02 | U_115.318 | NotIdentified | Not Identified | 0.000E+00 | 0.000E+00 | 0.000E+00 | 0.000E+00 | 8.379E-01 | 0.000E+00 | 4.645E+00 | 4.420E+02 | 4.470E+02 | 2.586E+00 | 0.000E+00 | False | |
| 1.153E+02 | U_115.334 | NotIdentified | Not Identified | 0.000E+00 | 0.000E+00 | 0.000E+00 | 0.000E+00 | 1.547E+00 | 0.000E+00 | 4.255E+00 | 4.830E+02 | 4.900E+02 | 4.775E+00 | 0.000E+00 | False | |
| 1.154E+02 | U_115.361 | NotIdentified | Not Identified | 0.000E+00 | 0.000E+00 | 0.000E+00 | 0.000E+00 | 1.520E+00 | 0.000E+00 | 1.804E+01 | 5.450E+02 | 5.630E+02 | 3.829E+00 | 0.000E+00 | False | |
| 1.154E+02 | U_115.367 | NotIdentified | Not Identified | 0.000E+00 | 0.000E+00 | 0.000E+00 | 0.000E+00 | 1.498E+00 | 0.000E+00 | 5.712E+00 | 5.680E+02 | 5.730E+02 | 4.623E+00 | 0.000E+00 | False | |
| 1.154E+02 | U_115.371 | NotIdentified | Not Identified | 0.000E+00 | 0.000E+00 | 0.000E+00 | 0.000E+00 | 1.349E+00 | 0.000E+00 | 4.023E+00 | 5.770E+02 | 5.820E+02 | 4.163E+00 | 0.000E+00 | False | |
| 1.154E+02 | U_115.387 | NotIdentified | Not Identified | 0.000E+00 | 0.000E+00 | 0.000E+00 | 0.000E+00 | 4.246E+01 | 0.000E+00 | 2.840E+01 | 5.840E+02 | 6.570E+02 | 1.311E+02 | 0.000E+00 | False | |
| 1.154E+02 | U_115.406 | NotIdentified | Not Identified | 0.000E+00 | 0.000E+00 | 0.000E+00 | 0.000E+00 | 1.884E+00 | 0.000E+00 | 2.003E+01 | 6.590E+02 | 6.830E+02 | 4.747E+00 | 0.000E+00 | False |
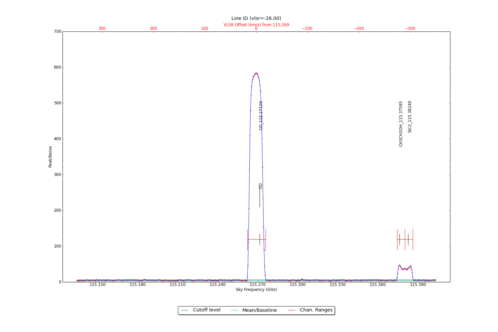
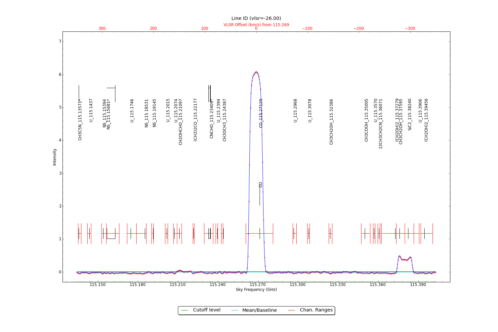
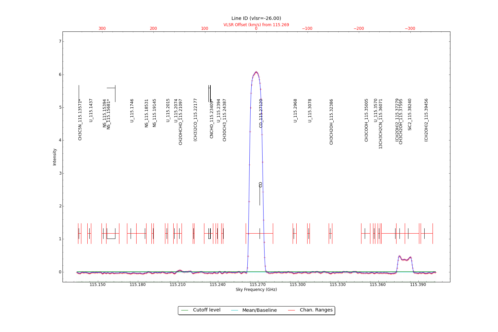
LineID_AT identifies spectral lines in an input datacube, using either one or more input spectra from CubeSpectrum_AT or CubeStats_AT.
Identified Spectral Lines
Identified Spectral Lines
| frequency | uid | formula | name | transition | velocity | El | Eu | linestrength | peakintensity | peakoffset | fwhm | startchan | endchan | peakrms | blend | force | [GHz] | [] | [] | [] | [] | [km/s] | [K] | [K] | [D^2] | [Jy/beam] | [km/s] | [km/s] | [] | [] | [] | [] | [] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1.151E+02 | CH3C5N_115.13571 | CH3C5N | Methylcyanodiacetylene | 74(0)-73(0),F=75-74 | -2.452E+01 | 2.017E+02 | 2.072E+02 | 4.374E+03 | -2.369E-02 | 1.481E+00 | 6.446E+00 | 2.000E+00 | 8.000E+00 | -5.203E+00 | 1.000E+00 | False |
| 1.151E+02 | U_115.1437 | U_115.1437 | Unknown | -2.600E+01 | 0.000E+00 | 0.000E+00 | 0.000E+00 | -2.391E-02 | 0.000E+00 | 9.224E+00 | 2.000E+01 | 2.700E+01 | -5.252E+00 | 0.000E+00 | False | |
| 1.152E+02 | NS_115.15394 | NS | Nitric sulfide | J=5/2-3/2,&Omega=1/2,F=7/2-5/2,l=e | -2.607E+01 | 4.422E+00 | 9.949E+00 | 1.048E+01 | -1.443E-02 | -6.528E-02 | 9.531E+00 | 4.700E+01 | 5.600E+01 | -3.169E+00 | 0.000E+00 | False |
| 1.152E+02 | NS_115.15681 | NS | Nitric sulfide | J=5/2-3/2,&Omega=1/2,F=5/2-3/2,l=e | -2.072E+01 | 4.424E+00 | 9.951E+00 | 6.606E+00 | -1.879E-02 | 5.280E+00 | 1.445E+01 | 5.600E+01 | 8.100E+01 | -4.127E+00 | 2.000E+00 | False |
| 1.152E+02 | NS_115.16298 | NS | Nitric sulfide | J=5/2-3/2,&Omega=1/2,F=3/2-1/2,l=e | 0.000E+00 | 4.425E+00 | 9.952E+00 | 3.931E+00 | 0.000E+00 | 0.000E+00 | 0.000E+00 | 5.600E+01 | 8.100E+01 | 0.000E+00 | 2.000E+00 | False |
| 1.152E+02 | U_115.1746 | U_115.1746 | Unknown | -2.600E+01 | 0.000E+00 | 0.000E+00 | 0.000E+00 | -2.443E-02 | 0.000E+00 | 1.386E+01 | 9.600E+01 | 1.140E+02 | -5.365E+00 | 0.000E+00 | False | |
| 1.152E+02 | NS_115.18531 | NS | Nitric sulfide | J=5/2-3/2,&Omega=1/2,F=3/2-3/2,l=e | -3.103E+01 | 4.424E+00 | 9.952E+00 | 1.257E+00 | -2.765E-02 | -5.031E+00 | 1.251E+01 | 1.140E+02 | 1.330E+02 | -6.074E+00 | 0.000E+00 | False |
| 1.152E+02 | NS_115.19145 | NS | Nitric sulfide | J=5/2-3/2,&Omega=1/2,F=5/2-5/2,l=e | -2.702E+01 | 4.422E+00 | 9.951E+00 | 1.257E+00 | -2.453E-02 | -1.018E+00 | 7.434E+00 | 1.430E+02 | 1.470E+02 | -5.389E+00 | 0.000E+00 | False |
| 1.152E+02 | U_115.2015 | U_115.2015 | Unknown | -2.600E+01 | 0.000E+00 | 0.000E+00 | 0.000E+00 | -2.289E-02 | 0.000E+00 | 1.022E+01 | 1.690E+02 | 1.740E+02 | -5.028E+00 | 0.000E+00 | False | |
| 1.152E+02 | U_115.2074 | U_115.2074 | Unknown | -2.600E+01 | 0.000E+00 | 0.000E+00 | 0.000E+00 | 1.955E-02 | 0.000E+00 | 4.249E+00 | 1.850E+02 | 1.920E+02 | 4.295E+00 | 0.000E+00 | False | |
| 1.152E+02 | CH2OHCHO_115.21097 | cis-CH2OHCHOv=0 | Glycolaldehyde | 23(5,19)-22(6,16) | -2.495E+01 | 1.643E+02 | 1.698E+02 | 1.473E+01 | 5.652E-02 | 1.051E+00 | 8.108E+00 | 1.920E+02 | 2.010E+02 | 1.241E+01 | 0.000E+00 | False |
| 1.152E+02 | (CH3)2CO_115.22177 | (CH3)2COv=0 | Acetone | 17(10,7)-16(13,4)EA | -2.589E+01 | 1.186E+02 | 1.241E+02 | 4.495E-01 | -2.753E-02 | 1.101E-01 | 5.268E+00 | 2.220E+02 | 2.250E+02 | -6.047E+00 | 0.000E+00 | False |
| 1.152E+02 | CNCHO_115.23280 | CNCHO | Cyanoformaldehyde | 6(2,5)-7(1,6),F=6-6 | 0.000E+00 | 1.623E+01 | 2.176E+01 | 4.333E-02 | 0.000E+00 | 0.000E+00 | 0.000E+00 | 2.440E+02 | 2.610E+02 | 0.000E+00 | 3.000E+00 | False |
| 1.152E+02 | CNCHO_115.23405 | CNCHO | Cyanoformaldehyde | 6(2,5)-7(1,6),F=5-6 | 0.000E+00 | 1.623E+01 | 2.176E+01 | 1.797E+00 | 0.000E+00 | 0.000E+00 | 0.000E+00 | 2.440E+02 | 2.610E+02 | 0.000E+00 | 3.000E+00 | False |
| 1.152E+02 | CNCHO_115.23409 | CNCHO | Cyanoformaldehyde | 6(2,5)-7(1,6) | -3.106E+01 | 1.623E+01 | 2.176E+01 | 6.371E+00 | -2.292E-02 | -5.061E+00 | 2.081E+01 | 2.440E+02 | 2.610E+02 | -5.035E+00 | 3.000E+00 | False |
| 1.152E+02 | CNCHO_115.23419 | CNCHO | Cyanoformaldehyde | 6(2,5)-7(1,6),F=6-7 | 0.000E+00 | 1.623E+01 | 2.176E+01 | 2.080E+00 | 0.000E+00 | 0.000E+00 | 0.000E+00 | 2.440E+02 | 2.610E+02 | 0.000E+00 | 3.000E+00 | False |
| 1.152E+02 | U_115.2394 | U_115.2394 | Unknown | -2.600E+01 | 0.000E+00 | 0.000E+00 | 0.000E+00 | -2.500E-02 | 0.000E+00 | 5.380E+00 | 2.660E+02 | 2.700E+02 | -5.491E+00 | 0.000E+00 | False | |
| 1.152E+02 | CH3OCH3_115.24387 | CH3OCH3 | Dimethyl ether | 32(8,24)-31(9,23)AE | -2.775E+01 | 5.654E+02 | 5.710E+02 | 1.436E+01 | -2.837E-02 | -1.753E+00 | 4.076E+00 | 2.770E+02 | 2.800E+02 | -6.232E+00 | 0.000E+00 | False |
| 1.153E+02 | CO_115.27120 | CO | Carbon Monoxide | 1-0 | -3.351E+01 | 0.000E+00 | 5.532E+00 | 1.212E-02 | 1.877E+02 | -7.513E+00 | 2.709E+01 | 3.240E+02 | 3.760E+02 | 5.796E+02 | 0.000E+00 | False |
| 1.153E+02 | U_115.2968 | U_115.2968 | Unknown | -2.600E+01 | 0.000E+00 | 0.000E+00 | 0.000E+00 | -2.572E-02 | 0.000E+00 | 2.031E+00 | 4.140E+02 | 4.210E+02 | -5.649E+00 | 0.000E+00 | False | |
| 1.153E+02 | U_115.3078 | U_115.3078 | Unknown | -2.600E+01 | 0.000E+00 | 0.000E+00 | 0.000E+00 | -2.614E-02 | 0.000E+00 | 2.031E+00 | 4.420E+02 | 4.460E+02 | -5.742E+00 | 0.000E+00 | False | |
| 1.153E+02 | CH3CH2OH_115.32386 | g-CH3CH2OH | gauche-Ethanol | 53(3,51)-53(2,51),vt=1-0 | -2.419E+01 | 1.230E+03 | 1.236E+03 | 3.236E+01 | -2.343E-02 | 1.810E+00 | 2.031E+00 | 4.830E+02 | 4.890E+02 | -5.146E+00 | 0.000E+00 | False |
| 1.154E+02 | CH3COOH_115.35005 | CH3COOHv=0 | Acetic Acid | 12(*,11)-12(*,12)-+v=0 | -1.917E+01 | 4.277E+01 | 4.831E+01 | 6.250E+00 | -2.166E-02 | 6.833E+00 | 2.030E+00 | 5.450E+02 | 5.620E+02 | -4.758E+00 | 0.000E+00 | False |
| 1.154E+02 | U_115.3570 | U_115.3570 | Unknown | -2.600E+01 | 0.000E+00 | 0.000E+00 | 0.000E+00 | -2.407E-02 | 0.000E+00 | 2.030E+00 | 5.680E+02 | 5.720E+02 | -5.287E+00 | 0.000E+00 | False | |
| 1.154E+02 | 13CH3CH2CN_115.36071 | 13CH3CH2CN | Ethyl Cyanide | 36(3,33)-36(2,34) | -2.657E+01 | 2.878E+02 | 2.933E+02 | 3.750E+01 | -2.372E-02 | -5.696E-01 | 2.030E+00 | 5.770E+02 | 5.810E+02 | -5.212E+00 | 0.000E+00 | False |
| 1.154E+02 | (CH2OH)2_115.37279 | g'Ga-(CH2OH)2 | Ethylene Glycol | 6(2,4)v=1-5(0,5)v=0 | -3.360E+01 | 7.233E+00 | 1.277E+01 | 2.105E+00 | -9.986E-03 | -7.601E+00 | 2.030E+00 | 5.840E+02 | 6.120E+02 | -2.194E+00 | 0.000E+00 | False |
| 1.154E+02 | CH3CH2OH_115.37595 | g-CH3CH2OH | gauche-Ethanol | 18(2,16)-18(1,17),vt=1-1 | -2.253E+01 | 2.063E+02 | 2.118E+02 | 9.947E-02 | 1.376E+01 | 3.472E+00 | 2.030E+00 | 6.120E+02 | 6.290E+02 | 4.246E+01 | 0.000E+00 | False |
| 1.154E+02 | SiC2_115.38240 | SiC2v=0 | Silicon Carbide | 5(0,5)-4(0,4) | -2.508E+01 | 1.123E+01 | 1.677E+01 | 2.846E+01 | 1.296E+01 | 9.211E-01 | 2.030E+00 | 6.290E+02 | 6.560E+02 | 4.001E+01 | 0.000E+00 | False |
| 1.154E+02 | U_115.3906 | U_115.3906 | Unknown | -2.600E+01 | 0.000E+00 | 0.000E+00 | 0.000E+00 | -8.811E-03 | 0.000E+00 | 2.030E+00 | 6.460E+02 | 6.560E+02 | -1.935E+00 | 0.000E+00 | False | |
| 1.154E+02 | (CH2OH)2_115.39456 | g'Ga-(CH2OH)2 | Ethylene Glycol | 31(5,26)v=1-31(5,27)v=0 | -2.318E+01 | 2.568E+02 | 2.623E+02 | 1.204E+02 | -1.449E-02 | 2.816E+00 | 2.029E+00 | 6.590E+02 | 6.820E+02 | -3.183E+00 | 0.000E+00 | False |
LineCube_AT makes one or more cutout cubes given a LineList. The purpose is to make small datacubes of the emission of individual spectral lines from a larger cube.
LineCube_AT created the following cubes
Parameters of Line Cubes
| Line Name | Start Channel | End Channel | Output Cube | [] | [int] | [int] | [] |
|---|---|---|---|---|
| CH3C5N_115.13571 | 0.000E+00 | 1.300E+01 | x.CH3C5N_115.13571/lc.im | |
| U_115.1437 | 1.500E+01 | 3.200E+01 | x.U_115.1437/lc.im | |
| NS_115.15394 | 4.200E+01 | 6.100E+01 | x.NS_115.15394/lc.im | |
| NS_115.15681 | 5.100E+01 | 8.600E+01 | x.NS_115.15681/lc.im | |
| U_115.1746 | 9.100E+01 | 1.190E+02 | x.U_115.1746/lc.im | |
| NS_115.18531 | 1.090E+02 | 1.380E+02 | x.NS_115.18531/lc.im | |
| NS_115.19145 | 1.380E+02 | 1.520E+02 | x.NS_115.19145/lc.im | |
| U_115.2015 | 1.640E+02 | 1.790E+02 | x.U_115.2015/lc.im | |
| U_115.2074 | 1.800E+02 | 1.970E+02 | x.U_115.2074/lc.im | |
| CH2OHCHO_115.21097 | 1.870E+02 | 2.060E+02 | x.CH2OHCHO_115.21097/lc.im | |
| (CH3)2CO_115.22177 | 2.170E+02 | 2.300E+02 | x.(CH3)2CO_115.22177/lc.im | |
| CNCHO_115.23409 | 2.390E+02 | 2.660E+02 | x.CNCHO_115.23409/lc.im | |
| U_115.2394 | 2.610E+02 | 2.750E+02 | x.U_115.2394/lc.im | |
| CH3OCH3_115.24387 | 2.720E+02 | 2.850E+02 | x.CH3OCH3_115.24387/lc.im | |
| CO_115.27120 | 3.190E+02 | 3.810E+02 | x.CO_115.27120/lc.im | |
| U_115.2968 | 4.090E+02 | 4.260E+02 | x.U_115.2968/lc.im | |
| U_115.3078 | 4.370E+02 | 4.510E+02 | x.U_115.3078/lc.im | |
| CH3CH2OH_115.32386 | 4.780E+02 | 4.940E+02 | x.CH3CH2OH_115.32386/lc.im | |
| CH3COOH_115.35005 | 5.400E+02 | 5.670E+02 | x.CH3COOH_115.35005/lc.im | |
| U_115.3570 | 5.630E+02 | 5.770E+02 | x.U_115.3570/lc.im | |
| 13CH3CH2CN_115.36071 | 5.720E+02 | 5.860E+02 | x.13CH3CH2CN_115.36071/lc.im | |
| (CH2OH)2_115.37279 | 5.790E+02 | 6.170E+02 | x.(CH2OH)2_115.37279/lc.im | |
| CH3CH2OH_115.37595 | 6.070E+02 | 6.340E+02 | x.CH3CH2OH_115.37595/lc.im | |
| SiC2_115.38240 | 6.240E+02 | 6.610E+02 | x.SiC2_115.38240/lc.im | |
| U_115.3906 | 6.410E+02 | 6.610E+02 | x.U_115.3906/lc.im | |
| (CH2OH)2_115.39456 | 6.540E+02 | 6.870E+02 | x.(CH2OH)2_115.39456/lc.im |
![Average Spectrum at centerbox[[48pix,48pix],[1pix,1pix]] Average Spectrum at centerbox[[48pix,48pix],[1pix,1pix]]](x.csp_0_thumb.png)
![Average Spectrum at centerbox[[09h47m57.257s,+13d16m42.91s],[1pix,1pix]] Average Spectrum at centerbox[[09h47m57.257s,+13d16m42.91s],[1pix,1pix]]](x.csp_1_thumb.png)

![Average Spectrum at centerbox[[13pix,86pix],[1pix,1pix]] Average Spectrum at centerbox[[13pix,86pix],[1pix,1pix]]](x.CH3C5N_115.13571/lc-@1.csp_0_thumb.png)

![Average Spectrum at centerbox[[9pix,82pix],[1pix,1pix]] Average Spectrum at centerbox[[9pix,82pix],[1pix,1pix]]](x.U_115.1437/lc-@2.csp_0_thumb.png)
![Average Spectrum at centerbox[[69pix,15pix],[1pix,1pix]] Average Spectrum at centerbox[[69pix,15pix],[1pix,1pix]]](x.NS_115.15394/lc-@3.csp_0_thumb.png)

![Average Spectrum at centerbox[[8pix,81pix],[1pix,1pix]] Average Spectrum at centerbox[[8pix,81pix],[1pix,1pix]]](x.NS_115.15681/lc-@4.csp_0_thumb.png)

![Average Spectrum at centerbox[[34pix,31pix],[1pix,1pix]] Average Spectrum at centerbox[[34pix,31pix],[1pix,1pix]]](x.U_115.1746/lc-@5.csp_0_thumb.png)

![Average Spectrum at centerbox[[9pix,82pix],[1pix,1pix]] Average Spectrum at centerbox[[9pix,82pix],[1pix,1pix]]](x.NS_115.18531/lc-@6.csp_0_thumb.png)
![Average Spectrum at centerbox[[78pix,81pix],[1pix,1pix]] Average Spectrum at centerbox[[78pix,81pix],[1pix,1pix]]](x.NS_115.19145/lc-@7.csp_0_thumb.png)
![Average Spectrum at centerbox[[38pix,28pix],[1pix,1pix]] Average Spectrum at centerbox[[38pix,28pix],[1pix,1pix]]](x.U_115.2015/lc-@8.csp_0_thumb.png)
![Average Spectrum at centerbox[[51pix,17pix],[1pix,1pix]] Average Spectrum at centerbox[[51pix,17pix],[1pix,1pix]]](x.U_115.2074/lc-@9.csp_0_thumb.png)

![Average Spectrum at centerbox[[50pix,47pix],[1pix,1pix]] Average Spectrum at centerbox[[50pix,47pix],[1pix,1pix]]](x.CH2OHCHO_115.21097/lc-@10.csp_0_thumb.png)
2CO_115.22177/lc-@10.mom_0_thumb.png)
2CO_115.22177/lc-@10.mom_1_thumb.png)
2CO_115.22177/lc-@10.mom_2_thumb.png)
![Average Spectrum at centerbox[[85pix,19pix],[1pix,1pix]] Average Spectrum at centerbox[[85pix,19pix],[1pix,1pix]]](x.(CH3)2CO_115.22177/lc-@11.csp_0_thumb.png)
![Average Spectrum at centerbox[[10pix,67pix],[1pix,1pix]] Average Spectrum at centerbox[[10pix,67pix],[1pix,1pix]]](x.CNCHO_115.23409/lc-@12.csp_0_thumb.png)
![Average Spectrum at centerbox[[34pix,37pix],[1pix,1pix]] Average Spectrum at centerbox[[34pix,37pix],[1pix,1pix]]](x.U_115.2394/lc-@13.csp_0_thumb.png)

![Average Spectrum at centerbox[[9pix,82pix],[1pix,1pix]] Average Spectrum at centerbox[[9pix,82pix],[1pix,1pix]]](x.CH3OCH3_115.24387/lc-@14.csp_0_thumb.png)
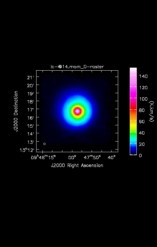
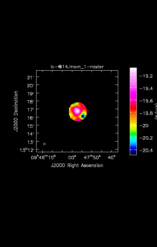
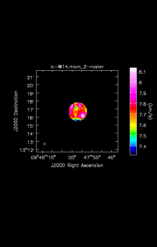
![Average Spectrum at centerbox[[48pix,48pix],[1pix,1pix]] Average Spectrum at centerbox[[48pix,48pix],[1pix,1pix]]](x.CO_115.27120/lc-@15.csp_0_thumb.png)
![Average Spectrum at centerbox[[86pix,55pix],[1pix,1pix]] Average Spectrum at centerbox[[86pix,55pix],[1pix,1pix]]](x.U_115.2968/lc-@16.csp_0_thumb.png)

![Average Spectrum at centerbox[[85pix,80pix],[1pix,1pix]] Average Spectrum at centerbox[[85pix,80pix],[1pix,1pix]]](x.U_115.3078/lc-@17.csp_0_thumb.png)
![Average Spectrum at centerbox[[12pix,13pix],[1pix,1pix]] Average Spectrum at centerbox[[12pix,13pix],[1pix,1pix]]](x.CH3CH2OH_115.32386/lc-@18.csp_0_thumb.png)
![Average Spectrum at centerbox[[45pix,27pix],[1pix,1pix]] Average Spectrum at centerbox[[45pix,27pix],[1pix,1pix]]](x.CH3COOH_115.35005/lc-@19.csp_0_thumb.png)
![Average Spectrum at centerbox[[15pix,31pix],[1pix,1pix]] Average Spectrum at centerbox[[15pix,31pix],[1pix,1pix]]](x.U_115.3570/lc-@20.csp_0_thumb.png)
![Average Spectrum at centerbox[[13pix,86pix],[1pix,1pix]] Average Spectrum at centerbox[[13pix,86pix],[1pix,1pix]]](x.13CH3CH2CN_115.36071/lc-@21.csp_0_thumb.png)
2_115.37279/lc-@21.mom_0_thumb.png)
2_115.37279/lc-@21.mom_1_thumb.png)
2_115.37279/lc-@21.mom_2_thumb.png)
![Average Spectrum at centerbox[[10pix,18pix],[1pix,1pix]] Average Spectrum at centerbox[[10pix,18pix],[1pix,1pix]]](x.(CH2OH)2_115.37279/lc-@22.csp_0_thumb.png)
![Average Spectrum at centerbox[[48pix,48pix],[1pix,1pix]] Average Spectrum at centerbox[[48pix,48pix],[1pix,1pix]]](x.CH3CH2OH_115.37595/lc-@23.csp_0_thumb.png)
![Average Spectrum at centerbox[[48pix,48pix],[1pix,1pix]] Average Spectrum at centerbox[[48pix,48pix],[1pix,1pix]]](x.SiC2_115.38240/lc-@24.csp_0_thumb.png)
![Average Spectrum at centerbox[[49pix,47pix],[1pix,1pix]] Average Spectrum at centerbox[[49pix,47pix],[1pix,1pix]]](x.U_115.3906/lc-@25.csp_0_thumb.png)
2_115.39456/lc-@25.mom_0_thumb.png)
2_115.39456/lc-@25.mom_1_thumb.png)
2_115.39456/lc-@25.mom_2_thumb.png)
![Average Spectrum at centerbox[[14pix,23pix],[1pix,1pix]] Average Spectrum at centerbox[[14pix,23pix],[1pix,1pix]]](x.(CH2OH)2_115.39456/lc-@26.csp_0_thumb.png)